| Entry Details |
| ID |
|
570 |
| Source Database |
|
UniProtKB/Swiss-Prot |
| UniProtKB/Swiss-Prot Accession Number |
|
P83120 (Created: 2001-11-02 Updated: 2008-12-16) |
| UniProtKB/Swiss-Prot Entry Name |
|
MTH_DROSI |
| Protein Name |
|
G-protein coupled receptor Mth |
| Gene |
|
mth |
| Organism Scientific |
|
Drosophila simulans |
| Organism Common |
|
Fruit fly |
| Lineage |
|
Eukaryota
Metazoa
Arthropoda
Hexapoda
Insecta
Pterygota
Neoptera
Endopterygota
Diptera
Brachycera
Muscomorpha
Ephydroidea
Drosophilidae
Drosophila
Sophophora
|
| Protein Length [aa] |
|
515 |
| Protein Mass [Da] |
|
59601 |
| Features |
|
| Type | Description | Status | Start | End | | signal peptide | | by similarity | 1 | 24 | | chain | G-protein coupled receptor Mth | | 25 | 515 | | disulfide bond | | by similarity | 29 | 83 | | disulfide bond | | by similarity | 85 | 90 | | disulfide bond | | by similarity | 94 | 188 | | disulfide bond | | by similarity | 95 | 106 | | disulfide bond | | by similarity | 150 | 209 | | transmembrane region | 1 | potential | 219 | 239 | | transmembrane region | 2 | potential | 249 | 269 | | transmembrane region | 3 | potential | 279 | 299 | | transmembrane region | 4 | potential | 321 | 341 | | transmembrane region | 5 | potential | 371 | 391 | | transmembrane region | 6 | potential | 425 | 445 | | transmembrane region | 7 | potential | 455 | 475 | | topological domain | Extracellular | potential | 25 | 218 | | topological domain | Cytoplasmic | potential | 240 | 248 | | topological domain | Extracellular | potential | 270 | 278 | | topological domain | Cytoplasmic | potential | 300 | 320 | | topological domain | Extracellular | potential | 342 | 370 | | topological domain | Cytoplasmic | potential | 392 | 424 | | topological domain | Extracellular | potential | 446 | 454 | | topological domain | Cytoplasmic | potential | 476 | 515 | | glycosylation site | N-linked (GlcNAc...) | by similarity | 45 | 45 | | glycosylation site | N-linked (GlcNAc...) | potential | 109 | 109 | | glycosylation site | N-linked (GlcNAc...) | by similarity | 123 | 123 | | glycosylation site | N-linked (GlcNAc...) | by similarity | 170 | 170 | | glycosylation site | N-linked (GlcNAc...) | potential | 198 | 198 | | glycosylation site | N-linked (GlcNAc...) | potential | 449 | 449 | | sequence variant | (in strain: HFL4.1S) | | 0 | 0 | | sequence variant | (in strain: HFL2.1S, VA7.1S, VA8.1S) | | 0 | 0 | | sequence variant | (in strain: GA2.1S) | | 0 | 0 | | sequence variant | (in strain: CT2.1S, CT5.1S, CT6.1S, VA6.1S) | | 0 | 0 | | sequence variant | (in strain: HFL2.1S, HFL4.1S, VA8.1S) | | 0 | 0 | | sequence variant | (in strain: GA2.1S, HFL2.1S, HFL4.1S, VA8.1S) | | 0 | 0 | | sequence variant | (in strain: GA2.1S, HFL2.1S, HFL4.1S) | | 0 | 0 | | sequence variant | (in strain: GA2.1S, GA4.1S, GA5.1S, HFL1.1S, HFL2.1S, HFL4.1S, VA7.1S, VA8.1S) | | 0 | 0 |
|
| SP Length |
|
24 |
| | ----+----1----+----2----+----3----+----4----+----5 |
| Signal Peptide | | MKTLFVLRISTVILVVLVIQKSYA |
|
Sequence | | MKTLFVLRISTVILVVLVIQKSYADILECDYFDTVDISAAQKLQNGSYLF
EGLLVPAILTGEYDFRILPDDSKQKVASHIRGCVCKLKPCVRFCCPHDHI
MDNGVCNDNMSEEELTELDPFLNVTLDDGSVSRRHFKNELIVQWDLPMPC
DGMFYLDNREEQDQYTLFENGTFFRHFDRVTLSKREYCLQHLTFADGNAT
SIRIAPHNCLIVPSITGQTVVMITSLICMVLTIAVYLFVKKLQNLHGKCF
ICYMVCLFMGYLFLLFDLWQISISFCKPAGVLGYFFVMAAFLWLSVISLH
LWNTFRGSSHKASRFFSEHRFLAYNTYAWGMSMVLTGITVLADNVVEDQD
WNPRVGEEGHCWIYTQAWSAMLYFYGPMVFLIAFNITMFILTARRIIGVK
KDIQNFAHRQERKQKLNSDKQTYTFFLRLFIIMGLSWSLEIGSYISQSNQ
TWANVFLVADYLNWSQGIIIFILFILKRSTLRLLQESIRGEGEEVDNSEE
EISLENTRFDRNVLS |
|
Original | | MKTLFVLRISTVILVVLVIQKSYADILECDYFDTVDISAAQKLQNGSYLF
EGLLVPAILTGEYDFRILPDDSKQKVASHIRGCVCKLKPCVRFCCPHDHI
MDNGVCNDNMSEEELTELDPFLNVTLDDGSVSRRHFKNELIVQWDLPMPC
DGMFYLDNREEQDQYTLFENGTFFRHFDRVTLSKREYCLQHLTFADGNAT
SIRIAPHNCLIVPSITGQTVVMITSLICMVLTIAVYLFVKKLQNLHGKCF
ICYMVCLFMGYLFLLFDLWQISISFCKPAGVLGYFFVMAAFLWLSVISLH
LWNTFRGSSHKASRFFSEHRFLAYNTYAWGMSMVLTGITVLADNVVEDQD
WNPRVGEEGHCWIYTQAWSAMLYFYGPMVFLIAFNITMFILTARRIIGVK
KDIQNFAHRQERKQKLNSDKQTYTFFLRLFIIMGLSWSLEIGSYISQSNQ
TWANVFLVADYLNWSQGIIIFILFILKRSTLRLLQESIRGEGEEVDNSEE
EISLENTRFDRNVLS |
| | ----+----1----+----2----+----3----+----4----+----5 |
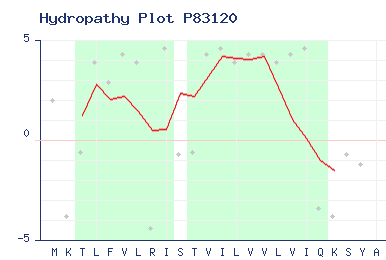
| Hydropathies |
|
 |
| |
|